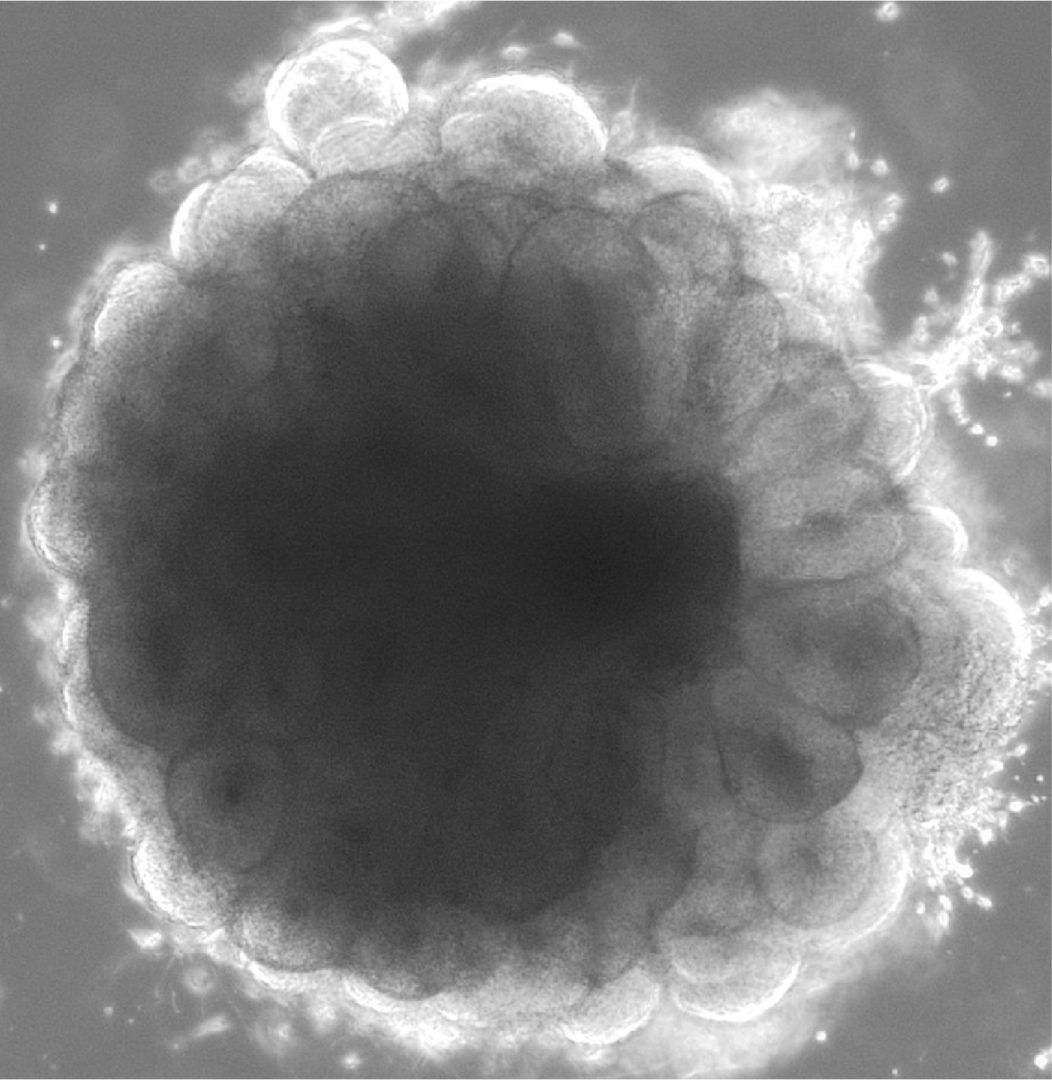
The mammalian cerebral cortex is the most complex biological structure of the animal kingdom and is in charge of our higher cognitive functions. Its development follows a sequence of genetically predefined molecular and cellular events that involve different types of neural precursors (NPs), including neural stem cells-like progenitors called Radial Glial Cells (RGCs), transient amplifying NPs (intermediate progenitors; IPs) and migratory neuroblasts (NBs). This process, generally referred to as neurogenesis, requires the activity of genes that are dynamically and specifically expressed across the different NP populations.
We investigate different aspects of the mechanisms that control the spatio-temporal expression of the developmental genes operating in cortical neurogenesis by using human cortical organoids (self-organized 3D cultures derived from embryonic or induced pluripotent stem cells that recapitulate embryonic cortex development and structure) as a model system. We combine functional genomics and chromatin epigenetic and interaction profiles analysis to address:
-
- The role of the non-coding genome in normal and pathological cortex development. Developmental genes are controlled by combinations of cis-regulatory elements (CREs) that can be located at long genomic distances from their target loci. Mutations in CREs associated with genes expressed in the embryonic cortex are linked with severe brain abnormalities, educational attainment and/or neuropsychiatric diseases. We use biochemical approaches, CRISPR/Cas9 genome editing and transgenesis approaches to identify and characterize the functions of CREs controlling neurodevelopmental gene expression, as well as to understand how mutations in these elements can impact the cortical neurogenesis process.
-
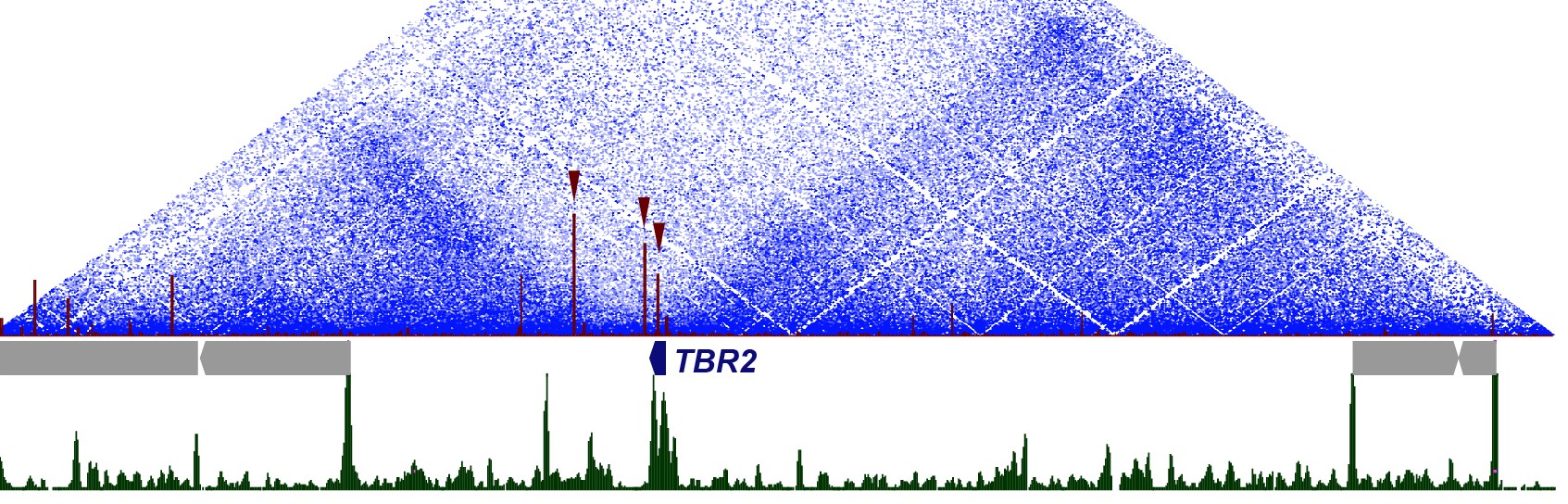
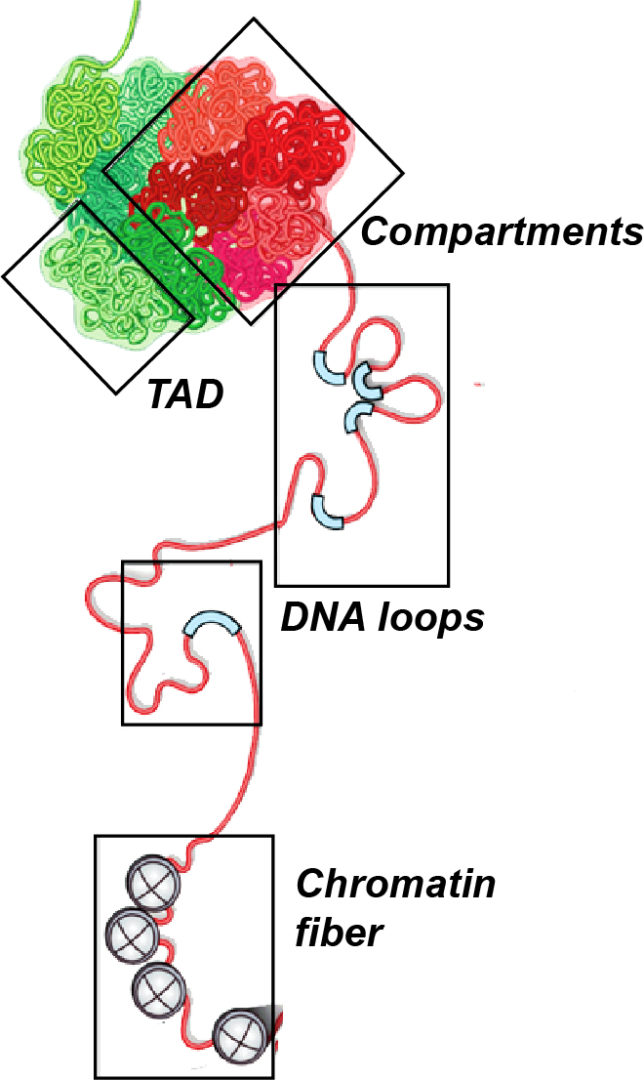
- The role of 3D chromatin architecture in neurodevelopmental gene regulation. The genomes of higher eukaryotes are hierarchically organized in 3D structures that contribute to gene expression regulation. Topologically Associated Domains (TADs), are megabase-scale structures in which interactions between internal DNA elements (eg: gene-enhancers) are favored compared to contacts with sequences located outside, constituting the fundamental unit in genome organization. TADs often coincide with and delimit the extent of gene regulatory landscapes (and can be identified across different tissues. We use functional genomics and chromatin conformation capture techniques to assess the 3D organization of the regulatory domains of neurodevelopmental genes and to understand how it contributes to their regulation.
-
- The mechanisms that control the activity of non-coding regulatory sequences. The accessibility of transcription factors and transcriptional machinery to the regulatory sequences controlling gene expression depends on the enzymatic activities that modulate the compaction and position of the nucleosome, as well as their histone composition and post-traductional modifications. We use biochemical and genome editing approaches to assess how epigenetic modifiers contribute to the control of cortical neurogenesis and how mutations affecting the activity of these proteins alter this process.
SÉLECTION DE PUBLICATIONS
- Rodríguez-Carballo E, Lopez-Delisle L, Willemin A, Beccari L, Gitto S, Mascrez M, and Duboule D. Chromatin topology and the timing of enhancer function at the HoxD locus. 2020. Proc Natl Acad Sci U S A. In press. BioRxiv (DOI: https://doi.org/10.1101/2020.07.12.199109)
- Vogg MC, Beccari L, Iglesias Ollé L, Rampon C, Vriz S and Galliot B. An evolutionary-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra. 2019. Nature Communications, 18;10(1):312. BioRxiv (DOI: https://doi.org/10.1101/265785).
- Beccari L*, Moris N*, Girgin M*, Turner DA, Baillie-Johnson P, Cossy AC, Lutolf M, Duboule D* and Martinez Arias A*. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. 2018. Nature, 562(7726):272-276.
- Schep R, Necsulea A, Rodríguez-Carballo E, Guerreiro I, Andrey G, Nguyen Huynh TH, Marcet V, Zákány J, Duboule D and Beccari L. 2016. Control of Hoxd gene transcription in the mammary bud by hijacking a preexisting regulatory landscape. Proc Natl Acad Sci U S A, 113:7720-9.
- Beccari L*, Yakushiji-Kaminatsui N*, Woltering JM*, Necsulea A, Lonfat N, Rodríguez-Carballo E, Mascrez B, Yamamoto S, Kuroiwa A and Duboule D. 2016. A role for HOX13 proteins in the regulatory switch between TADs at the HoxD locus. Genes Dev., 30(10):1172-86. *equal contribution.
- Beccari L#, Marco-Ferreres R, Tabanera N, Manfredi A, Souren M, Wittbrodt B, Conte I, Wittbrodt J and Bovolenta P#. 2015. A trans-regulatory code for the forebrain expression of Six3.2 in the medaka fish. J Biol Chem, 290:26927-42. #: Co-corresponding authors.
- Gómez-Marín C, Tena JJ, Acemel RD, López-Mayorga M, Naranjo S, de la Calle-Mustienes E, Maeso I, Beccari L, Aneas I, Vielmas E, Bovolenta P, Nobrega MA, Carvajal J and Gómez-Skarmeta JL. 2015. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci USA, 112: 7542-7.
- Ferri A, Favaro R*, Beccari, L*, Bertolini J, Tosetti V, Verzeroli C, Mercurio S, La Regina F, Ottolenghi S, Bovolenta P and Nicolis SK. 2013. Sox2 is required for embryonic development of the ventral telencephalon through the activation of the ventral determinants Nkx2.1 and Shh. Development, 140: 1250-61. *equal contribution.
- Beccari L, Marco-Ferreres R and Bovolenta P. 2013. The logic of gene regulatory networks in early vertebrate forebrain patterning. Mech Dev., 130: 95 -111.
- Beccari L, Conte I*, Cisneros E* and Bovolenta P. 2012. Differential Six3.2 activity patterns the forebrain through Sox2 dependent and independent mechanisms. Development 139: 151-64. *equal contribution.
FINANCEMENTS



Adresse
Faculté de Médecine
3ème étage, Aile B
8 Avenue Rockefeller
F-69008 Lyon


